LD Motif Finder locates ancient hidden protein patterns
06 January, 2020
An iterative machine learning approach has identified elusive 800 million-year-old amino acid patterns that are responsible for facilitating protein interactions.
Leucine-aspartic acid (LD) motifs are short amino acid sequences embedded within some proteins to link them to cellular molecules that control cell adhesion, motility and survival. They are known to also play a role in cancer cell spreading and in cardiovascular and infectious diseases. LD motifs were first revealed in 1996 in a family of proteins called paxillin. Only three other LD motif-containing proteins have been discovered since then, and scientists do not know the importance of LD motifs or how many other types of proteins contain them.
Click here to read the full story
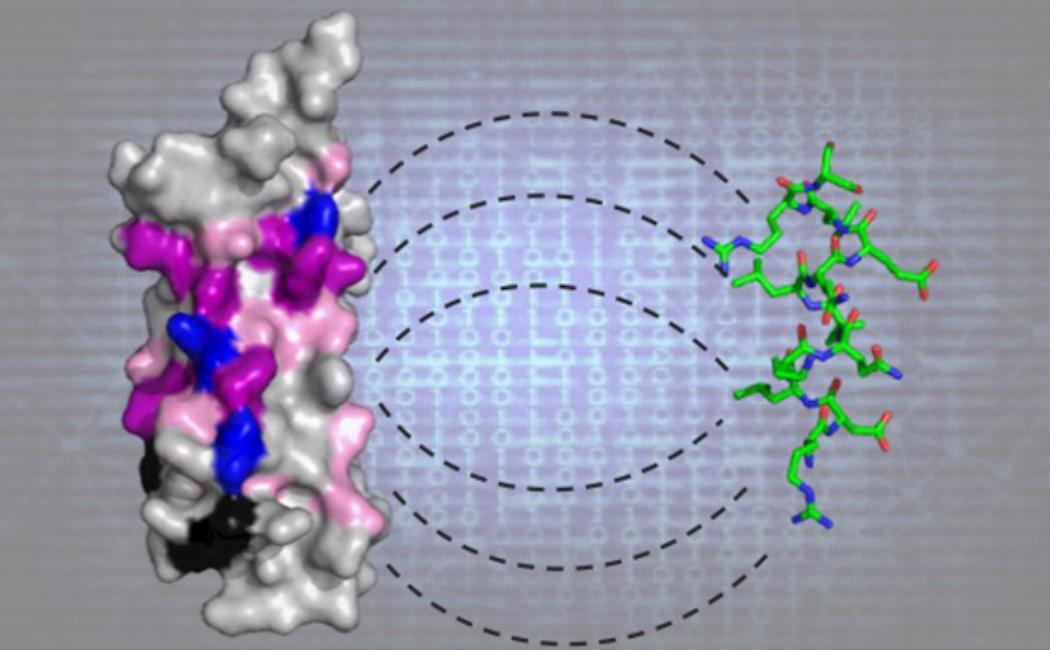
Image: The researchers used AI methods to predict the interaction (shown by the dashed lines) of short, linear protein segments (green stick model) with their target proteins (molecular representation on the left). Predictions were experimentally confirmed as shown by the imprint of the ligand on the surface of the target protein (blue, magenta and pink colored surfaces).
© 2019 Rayan Naser