Uncovering hidden mitochondrial mutations in single cells
13 April, 2023
A high-throughput single-cell single-mitochondrial genome sequencing technology known as iMiGseq has provided new insights into mutations of mitochondrial DNA (mtDNA) and offers a platform for assessing mtDNA editing strategies and genetic diagnosis of embryos prior to their implantation.
An international team of researchers, led by KAUST stem cell biologist Mo Li, has now quantitatively depicted the genetic maps of mtDNA in single human oocytes (immature eggs) and blastoids (stem cell-based synthetic embryos)article " id="return-reference-1" href="https://discovery.kaust.edu.sa/en/article/21015/uncovering-hidden-mitochondrial-mutations-in-single-cells/#reference-1">[1]. This has revealed molecular features of rare mtDNA mutations that cause maternally inherited diseases.
Mitochondria, the “powerhouses” of cells, play a crucial role in cellular communication and metabolism. Human mtDNA is a circular genome containing 37 genes, encoding 13 proteins and a noncoding D-loop region. Heteroplasmic mutations, inherited from egg cells, can cause congenital diseases, like maternally inherited Leigh syndrome, and are associated with late-onset complex diseases.
Click here to read the full story.
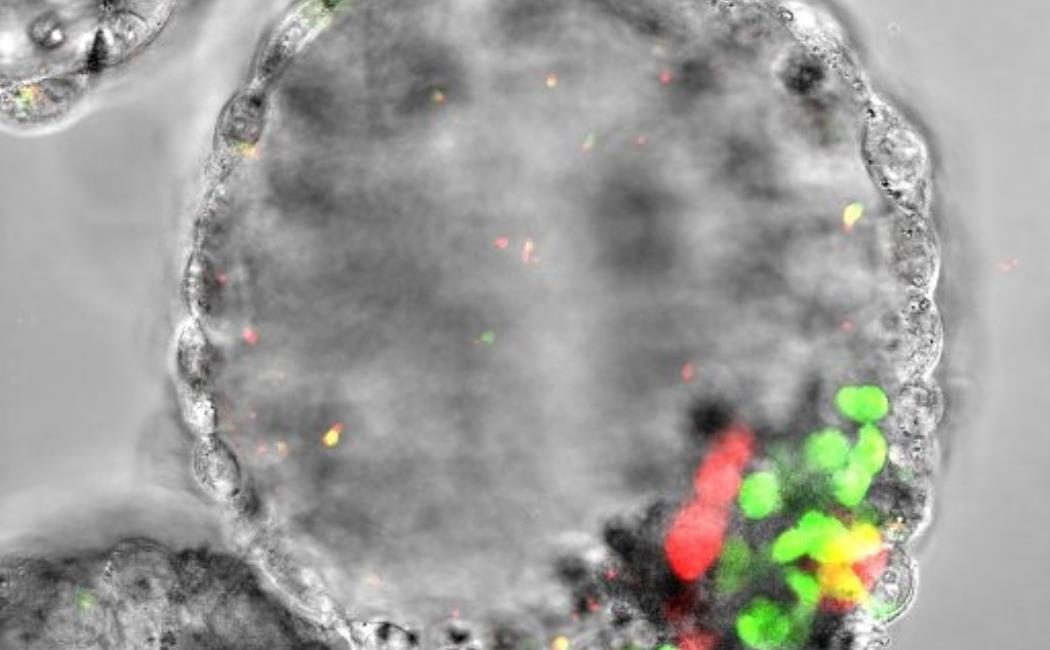
Image: A human blastocyst-like synthetic embryo called blastoid showing the presence of an enveloping layer of extra-embryonic cells, a blastocoel-like cavity, epiblast cells (green, giving rise to the future embryo) and hypoblast cells (red, giving rise to the future amnion). iMiGSeq was used to sequence mtDNA in a single blastoid to model the dynamics of mtDNA mutations during human embryogenesis. © 2023 KAUST; Mo Li.